Exploratory Work on the Predictive Clinical Neuroscience (PCN) Toolkit
By Andjela Dimitrijevic
Published on July 29, 2022
| Last updated on
August 7, 2022

Project definition
Background
The PCN toolkit aims at providing diffferent tools of normative modelling for understanding psychiatric disorders at the individual level including:
- Data selection,
- Data preparation,
- Algorithm & modelling,
- Evaluation & interpretation
I was interesting in how to use this tool because it overlaps with my PhD work on trying to find a neuroimaging atlas for pediatric population. Hence, I wanted to see if it would be useful with the data I have.
Objectives
The aim of this project will be decomposed into three sub-objectives:
- Start off with the available tutorial on the PCN toolkit documentation to get familiar to it
- Use another set of data if possible to follow through the available steps that this toolkit offers
- Adapt some functionalities which were found to be less intuitive to work with
- Learn some new tools throughout this work
All of these three sub-objectives will be documented to be able to be reproduced in later or other exploratory works.
Tools
This project relied on numerous tools throughout its creation:
- PCN Toolkit and their GitHub page to explore their tutorials
- Google Colab to follow through some of the tutorials
- Bash Terminal constantly when moving files or using Git
- High performance computing (HPC) through accessing the BrainHack cloud
- Markdown to structure the tutorial guides
- Git and GitHub for version control and making the project reproducible

Data
In the end, this project does not use data a lot because guides for using some neuroimaging tools are presented instead. However, FreeSurfer was used on two subjects of the publicly available longitudinal Calgary Preschool Dataset.
Results: Deliverables of this project
At the end of this BrainHack school, these files will be made available:
- A GitHub page containing all the modifications that were made to the PCN-toolkit tutorials
- A markdown file on where to start with the PCN-toolkit tutorials and another on how to format the data adequately all in the PCNTutorial_steps directory on GitHub
- A markdown file on getting connected to the BrainHack Cloud to access HPC clusters as well as one on testing out the Neurodesktop container all in the BrainHackCloud_steps directory on GitHub
Progress overview
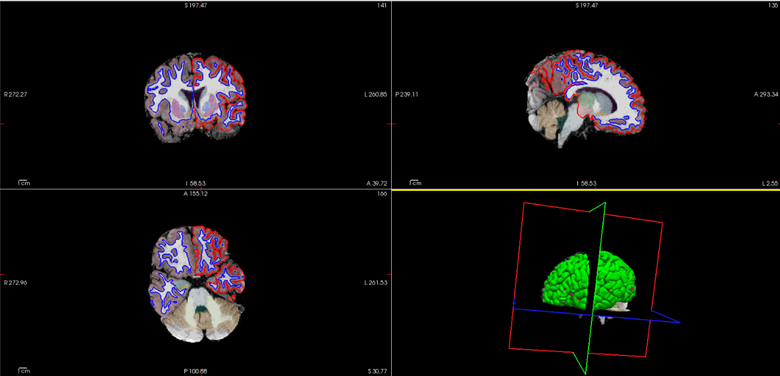
At first, I thought using the Calgary Preschool Dataset would have been enough to do the analysis with the PCN-toolkit. However, this dataset seems too small (only 64 subjects) for that type of analysis. Furthermore, it was lacking cortical thickness maps which is an essential feature to be analyzed. Hence, that's why I started connecting to the HPC during the last week of the BrainHack school to be able to run FreeSurfer recon-all function on all subjects. This too came with its specific challenges. First, getting connected to the BrainHack cloud from my own computer did not work directly. Second, it took quite long to run the wanted FreeSurfer command on the NeuroDesk test server on one subject. Also, FreeSurfer bases its cortical thickness maps on previous registration tasks using adult templates (by default the Talairach atlas). This can incorporate biases and render the results to be faulty as shown on the two figures below. Indeed, some regions are not well delineated whether it be for the white matter and gray matter segmentation on Fig. 1 or the wrong specific parcellations shown by the green rectangle on the second figure.
 Fig. 1 Example of white matter segmentation on a subject
Fig. 1 Example of white matter segmentation on a subject
 Fig. 2 Parcellations of the right and left hemisphere pial surface for cortical analyses
Fig. 2 Parcellations of the right and left hemisphere pial surface for cortical analyses
The final presentation on this project is avaible via this link for further details.
Detailed actions
Reported GitHub issue to alert the PCN-toolkit tutorial writers
The BLR_protocol tutorial is more easily ran locally and does not work because of the older python version available on Google Colab. The solution of running it locally has all been explained in the where_to_start md file. To consult the reported issue click here
Accessed the neurodesktop via the BrainHack Cloud successfully
The neurodesktop test server is great and easily accessible through the browser with various neuroimaging tools. Their test server is available via this link without any installation required. Also, installing and accessing this neurodesktop container through the BrainHack cloud was succesful. However, an important note is that running the NeuroDesk container via Docker should first be done on a virtual machine and NOT directly on the cloud.
Obtained recon-all FreeSurfer results for one subject
This was ran directly on the test server which is an interesting tool easily usable by everyone. The results could not be loaded as a zip file in the main GitHub repository for this project because of their size. However, this took approximately 7 hours to run because it was done on the available resources on the test server. Hence, using BrainHack cloud resources to parallelize everything is suggested.
Future Work
Some future steps which will be done to follow through this project are presented below:
- Aggregating multiple pediatric datasets to have more subjects and also different sites to analyze with the PCN-Toolkit
- Submitting a job to the slurm scheduler to obtain cortical thickness maps of the data, but with an appropriate atlas
- Uploading this newly modified data via DataLad tools to make it more reproducible
Conclusion and acknowledgement
To conclude, the initial objectives of this project have been slightly modified. However, I am still glab with all the new essential tools I have learned throughout the weeks during this BrainHack school. I would like to thank all the instructors, but specifically the ones from the group-analysis pod which made this experience smoother! 🎆