Workflow of resting state connectivity from a raw dataset to longitudinal visualization
By Kevin Jamey
Published on August 7, 2022
| Last updated on
September 27, 2022

Project definition
ABOUT ME
My research interests are on new technologies for music-based cognitive training in children with neurodevelopmental disorders. I completed my MSc. on music perception abilities in children on the Autism Spectrum (AS) at the University of Montreal under the supervision of Krista L. Hyde. My doctoral thesis is supervised by Simone Dalla Bella and examines the neural correlates of a tablet-based rhythmic training game (RhythmWorkers) on executive functioning and speech in children on the AS. In my free time, I am a singer-songwriter, music producer/live act and I love to create powerful musical experiences where lyrics are inflected for rhythm and melody.
PROJECT SUMMARY
Main objectives
- Provide a full neuroimaging workflow from preprocessing of raw data to visualisation of results
- To explore longitudinal analysis between two treatments in this dataset
- To analyse resting-state networks linked to joint attention (dorsal attentional, visual, DMN)
Personal objectives
- To learn the full workflow of neuroimaging from converting raw data to preprocessing to data visualisation
- To set myself up with open science tools for neuroimaging work in the future
- To get familiar with the best practices of shareable open science kits and software
Tools
- Git and Github for sharing project
- dcm2bids for converting raw data into NiFTY and a BIDS structure
- fMRIPrep for data preprocessing
- Alliance Canada for computing pre-processing
- Python Packages matplotlib, nilearn, plotly
Data
Dataset was collected during my masters in 2016 at the MNI. I scanned ~50 participants with autism before and after either 12 weeks of music therapy or arts and crafts therapy. The data involves a weighted T1 scan and resting-state fMRI. A previous publication on this data set can be found here.
Project deliverables
- project workflow detailed in my GitHub repository
- the bash code I used to run BIDS conversion and fMRIprep
- a markdown file introducing the project, highlighting the results and a discussion with recommendations
Results
Progress overview
The project was workflow was set-up for a single participant over the course of the brainhack school. It will be adapted in the future in order to run multiple participants and do group seed-based resting state fMRI.
Tools I learned during this project
- Project-Management I learnt how to set-up the framework for making a shareable neuroimaging project using open science rules.
- Github workflow- I learnt to use GitHub in order to share my project openly and invite people to collaborate on it.
- Project content Here you will find all the key steps necessary to move from a raw imaging dataset to some cool visualisations.
Results
Deliverable 1: report template
You are currently reading the report template! I will let you judge whether it is useful or not. If you think there is something that could be improved, please do not hesitate to open an issue here and let me know.
Deliverable 2: Visualisation using Nilearn
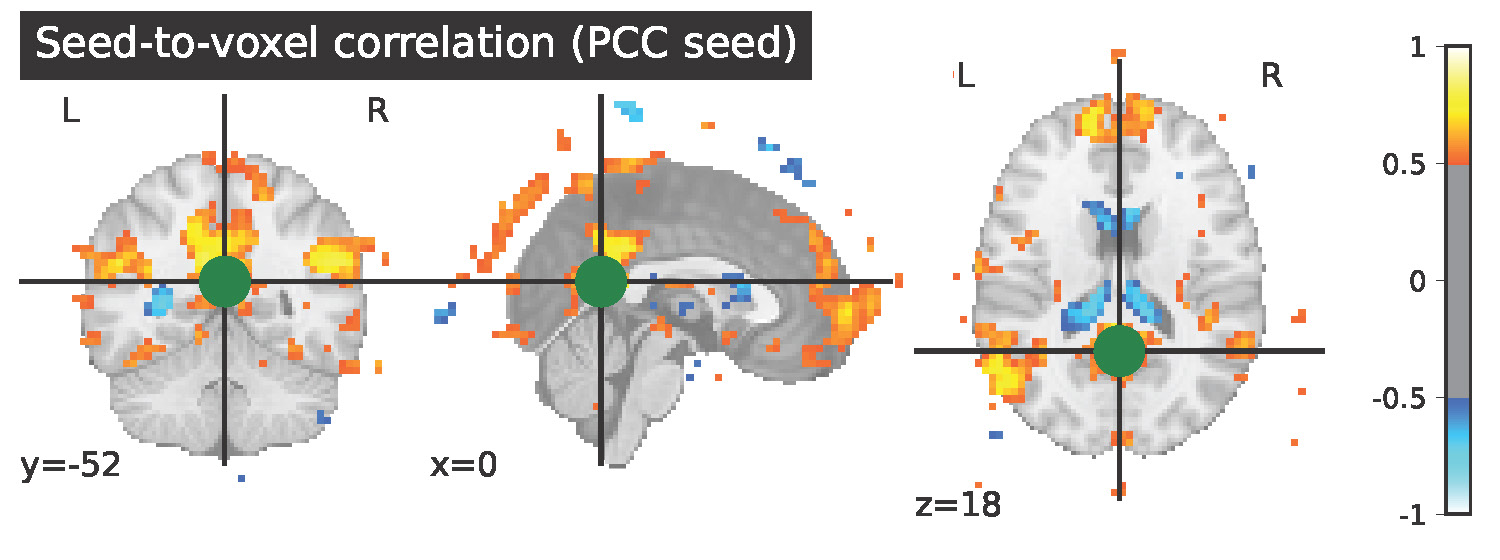
After successfully convertion the raw data of this participant to BIDS and NiFTY, I preprocessed his data using fmriprep. You can see details about this in my slides. I looked at a simple connectome before and after a music intervention for a child diagnosed on the austim spectrum. This is simply for illustration purposes since a large amount of participants are requited to make a meaningful connectome. Then visualized seed-based connectivity of the Posterior Cingulate Cortex before and after music therapy in this subject. You can see the visual results of my project here. Behaviorally we found differences after the music therapy in joint attention. In this single subject's seed-based conectivity there seems to be more connectivity in the medial prefrontal cortex, the visual cortex, and tempo-parietal network, which would be in line with improvement in joint attention. In order to further develop these findings for the experimental group, a seed-based connectivity for the entire experimental and control treatment will be conducted.
Conclusion and acknowledgement
I hope you find this projet useful in outlining a standard shareable workflow from imgages fresh out of the scanner to tangible visualisations of resting-state connectivity. Many thakns to the brainhack school instructors and students who helped make this prossible in such a short amount of time. I look forward to devling deeper into the foundations that are laid here during my PhD.